Scientists Develop Novel SERS Biosensors to Identify Infectiousness of SARS-Cov-2
Coronavirus disease 2019 (COVID-19), caused by SARS-CoV-2, is a serious threat to human health. Since July 2020, identification of SARS-CoV-2 virions on cold chain items has been frequently reported in China and other countries, causing great concerns on relevant virus transmission. It can be attributed to the fact that SARS-COV-2 can survive more than 3 weeks below 0℃ during cold chain transportation.
It is important to note that a portion of SARS-COV-2 virions in the colder environment are lysed, inactive viruses. But the RNA of lysed SARS-CoV-2 might remain and easily be detected by the universally applied polymerase chain reaction (PCR) method, resulting in the misdiagnosis of infectiousness for SARS-CoV-2, causing unnecessary social panic.
Therefore, it is much more meaningful to determine whether the virus is infectious as identified on contaminated items in various environments. However, exploiting a rapid and highly sensitive detection method that can identify lysed, noninfectious SARS-CoV-2 virions to rule out the contamination of infectious SARS-CoV-2 virus is a key scientific challenge.
Surface-enhanced Raman scattering (SERS), a single-molecule spectral detection technology, is currently expanding its promising applications from environmental science and food safety toward the biosensing field due to its advantages of ultra-high sensitivity, non-destructiveness, excellent repeatability, and accuracy.
Recently, the scientists from Shanghai Institute of Ceramics Chinese Academy of Sciences developed one novel SnS2 semiconductor-based SERS biosensors, and it exhibited an extremely low limit of detection of 10-13 M for methylene blue, which is one of the highest sensitivities among the reported pure semiconductor-based SERS substrates. Such ultra-high SERS sensitivity originated from the synergistic enhancements of the molecular enrichment caused by capillary effect and the charge transfer chemical enhancement boosted by the lattice strain and sulfur vacancies.
The novel two-step SERS diagnostic route based on the ultra-sensitive SnS2 substrate was presented to diagnose the infectiousness of SARS-CoV-2 through the identification standard of SERS signals for SARS-CoV-2 S protein and RNA, which could accurately identify non-infectious lysed SARS-CoV-2 virions in actual environments, whereas the current PCR methods cannot.
As a result, various physical forms of SARS-CoV-2 were able to be sensitively detected on SnS2 microspheres, and the identification standard of SARS-CoV-2 RNA and S protein was established by PCA methods. Moreover, based on the advanced machine learning method of SVM, non-infectious lysed SARS-CoV-2 was successfully distinguished, which paved a new path for identifying the infectiousness of SARS-CoV-2 virions and is of significance to avoid misdiagnosing infectious SARS-CoV-2 in actual environments.
Furthermore, it is worthwhile to note that recovery of SARS-CoV-2 in viral culture is currently the only approach to confirm the presence of replication-competent virus. However, the viral culture method suffers from the defects of long culture time and complicated experimental operation. Meaningfully, the aforementioned two-step SERS detection method can be extended to rapidly diagnose SARS-CoV-2 infectivity on site in some contaminated patient gathering places such as hospitals or at the Centers for Disease Control and Prevention, exhibiting vital timeliness in patient management that the viral culture method does not have.
Relevant results have been published in Matter entitled “Identifying infectiousness of SARS-CoV-2 by ultra-sensitive SnS2 SERS biosensors with capillary effect” (https://doi.org/10.1016/j.matt.2021.11.028).
This work was supported by the National Key Research and Development Program of Anhui Province, and National Natural Science Foundation of China.
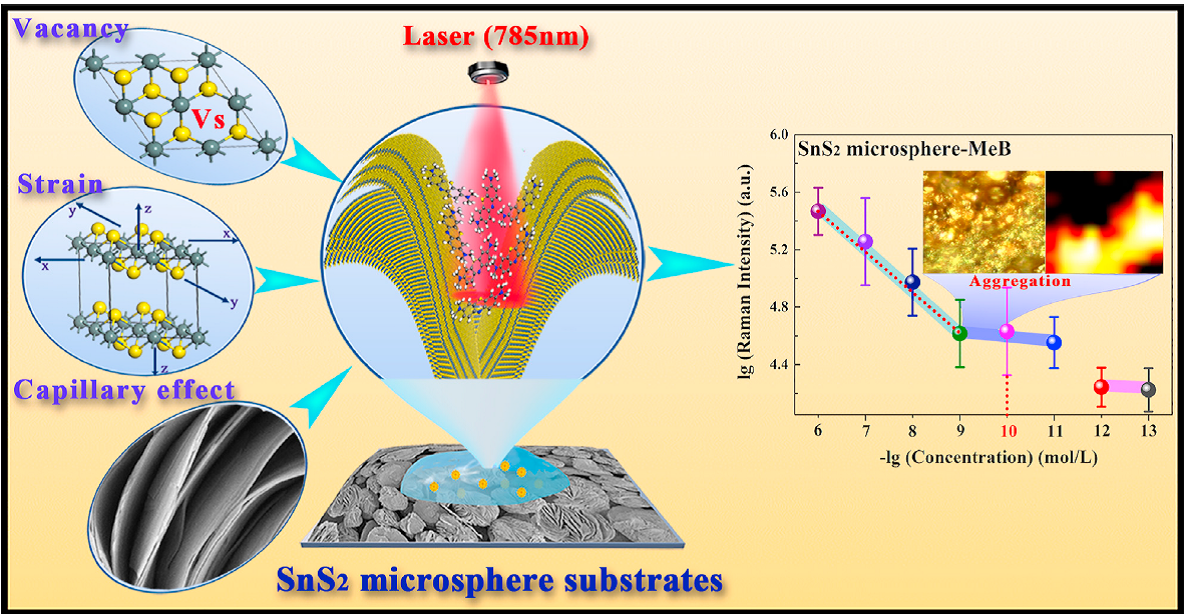
Figure 1. Schematic diagram of SnS2 microsphere substrates design and the ultra-sensitive SERS performance
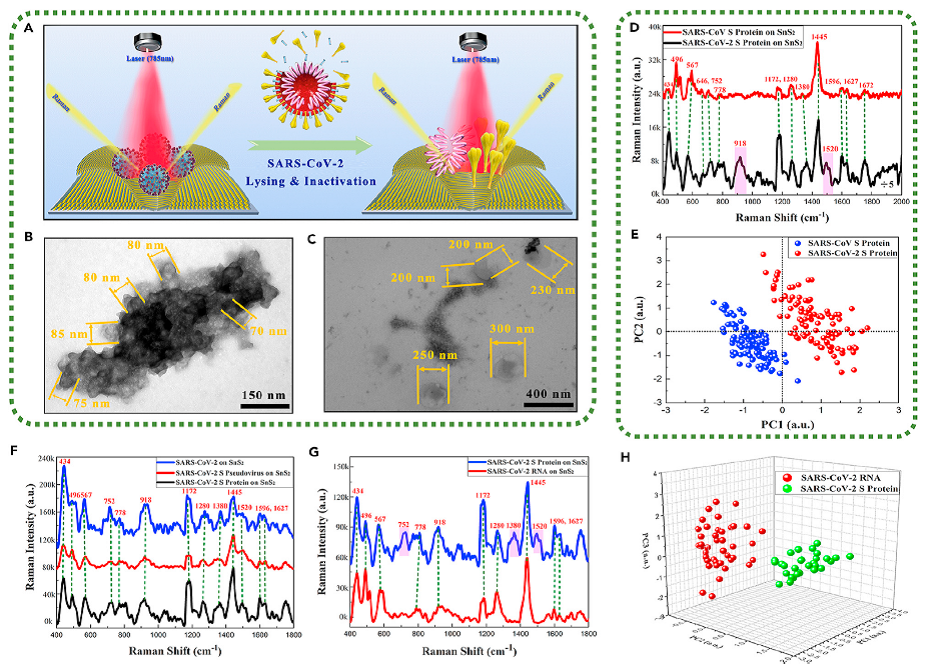
Figure 2. Application of SnS2 microspheres for detecting various physical forms of SARS-CoV-2
(A) Schematic diagram of identifying the lysed SARS-CoV-2.
(B) TEM images of SARS-CoV-2 with complete viral structure.
(C) TEM images of the lysed SARS-CoV-2.
(D) Raman spectra of SARS-CoV-2 S protein and SARS-CoV S protein.
(E) The key features of SERS patterns to classify the SARS-CoV-2 S protein and SARS-CoV S protein.
(F) Raman spectra of physical forms of SARS-CoV-2 including SARS-CoV-2, SARS-CoV-2 S pseudovirus, and SARS-CoV-2 S protein.
(G) Raman spectra of SARS-CoV-2 RNA and SARS-CoV-2 S protein.
(H) The key features of SERS patterns to classify the RNA and S protein of SARS-CoV-2.
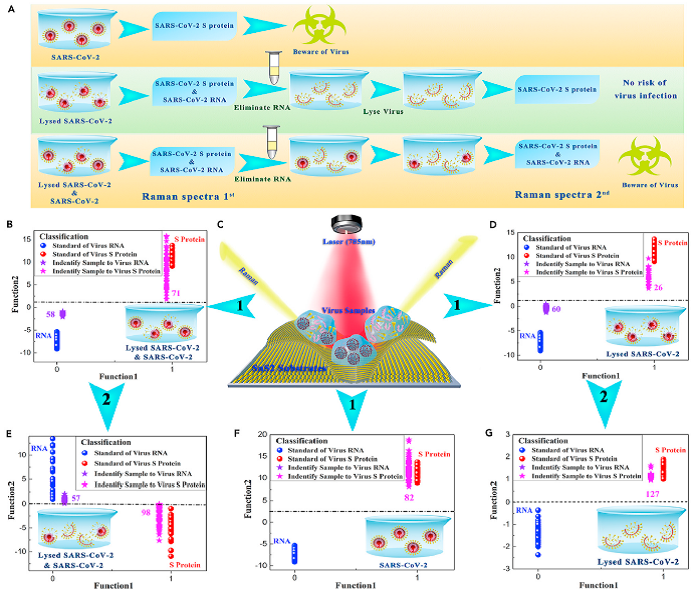
Figure 3. Application of SnS2 microspheres for diagnosing the infectiousness of SARS-CoV-2
(A) Experimental procedure for diagnosing the infectiousness of SARS-CoV-2.
(B) SVM analysis results to identify the mixture of the SARS-CoV-2 with complete viral structure and the lysed SARS-CoV-2.
(C) Raman scattering diagram of three contamination situations of the novel coronavirus based on SnS2 substrates.
(D) SVM analysis results to identify the lysed SARS-CoV-2.
(E) SVM analysis results to identify the mixture of the SARS-CoV-2 with complete viral structure and the lysed SARS-CoV-2 after eliminating RNA and relysing virus samples.
(F) SVM analysis results to identify the SARS-CoV-2 with complete viral structure.
(G) SVM analysis results to identify the lysed SARS-CoV-2 after eliminating RNA and re-lysing virus samples.
Reference:
https://www.cell.com/matter/fulltext/S2590-2385(21)00621-4
Contact:
Prof. YANG Yong
Shanghai Institute of Ceramics, CAS
Email: yangyong@mail.sic.ac.cn



